108985
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

基因表达谱分析显示了糖尿病周围神经病变的候选生物标志物和可能的分子机制
Authors Zhou H, Zhang W
Received 19 March 2019
Accepted for publication 4 June 2019
Published 23 July 2019 Volume 2019:12 Pages 1213—1223
DOI https://doi.org/10.2147/DMSO.S209118
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Amy Norman
Peer reviewer comments 2
Editor who approved publication: Professor Ming-Hui Zou
Purpose: To investigate the molecular mechanism and search for candidate biomarkers in the gene expression profile of patients with diabetic peripheral neuropathy (DPN).
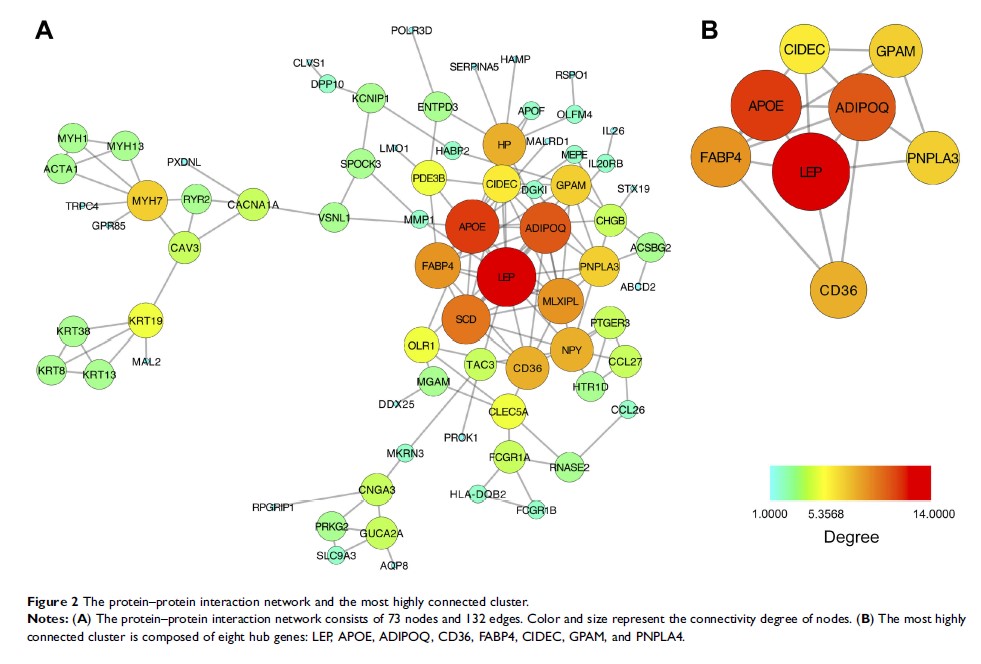
Methods: Differentially expressed genes (DEGs) of progressive vs non-progressive DPN patients in dataset GSE24290 were screened. Functional enrichment analysis was conducted, and hub genes were extracted from the protein–protein interaction network. The expression level of hub genes in serum samples in another dataset GSE95849 was obtained, followed by the ROC curve analysis.
Results: A total of 352 DEGs were obtained from dataset GSE24290. They were involved in 14 gene ontology terms and 10 Kyoto Encyclopedia of Genes and Genomes pathways, mainly related to lipid metabolism. Eight hub genes (LEP, APOE, ADIPOQ, FABP4, CD36, GPAM, CIDEC, and PNPLA4) were revealed, and their expression level was obtained in dataset GSE95849. The receiver operatingcharacteristic curve analysis indicated that CIDEC (AUC=1), APOE (AUC=0.833), CD36 (AUC=0.803), and PNPLA4 (AUC=0.861) might be candidate serum biomarkers of DPN.
Conclusion: Lipid metabolism of Schwann cells might be inhibited in progressive DPN. CIDEC, APOE, CD36, and PNPLA4 might be potential predictive biomarkers in the early DPN diagnosis of patients with DM.
Keywords: differentially expressed genes, functional enrichment analysis, demyelination, lipid metabolism, diabetic peripheral neuropathy