9 7 8 1 6
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.3 Breast Cancer (Dove Med Press)
- 3.4 Clin Epidemiol
- 2.5 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.5 Clin Interv Aging
- 4.7 Drug Des Dev Ther
- 2.7 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.5 Int J Women's Health
- 2.5 Neuropsych Dis Treat
- 2.7 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.3 Ther Clin Risk Manag
- 2.5 J Pain Res
- 2.8 Diabet Metab Synd Ob
- 2.8 Psychol Res Behav Ma
- 3.0 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.7 Risk Manag Healthc Policy
- 4.2 J Inflamm Res
- 2.1 Int J Gen Med
- 4.2 J Hepatocell Carcinoma
- 3.7 J Asthma Allergy
- 1.9 Clin Cosmet Investig Dermatol
- 2.7 J Multidiscip Healthc

Linc01194 在结肠直肠癌中作为致癌基因,并且与较差的存活率相关
Authors Wang X, Liu Z, Tong H, Peng H, Xian Z, Li L, Hu B, Xie S
Received 29 September 2018
Accepted for publication 31 December 2018
Published 22 March 2019 Volume 2019:11 Pages 2349—2362
DOI https://doi.org/10.2147/CMAR.S189189
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Andrew Yee
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Background: The
incidence of colorectal cancer ranks among the top three malignant tumors,
attributing to more than 50,000 deaths in the United States every year.
Survival rate is directly correlated with TNM stage at diagnosis, and
identifying the molecules involved in the cancer development process will
provide directions to better investigate the mechanisms of colorectal cancer.
Materials and methods: Bioinformatics
analysis of differentially expressed long noncoding RNAs (lncRNAs), survival
analysis, cell proliferation assay, migration assay, and Western blot analysis
were performed.
Results: Fifty-one
lncRNAs were identified between the early stage and late-stage groups. In the
survival analysis, we found that Linc01194 is correlated with poor survival of
colon cancer patients. In addition, by suppressing the expression of Linc01194
in colon cancer cell lines, cell proliferation and migration were inhibited.
Western blot showed that N-cadherin and vimentin were downregulated, whereas
E-cadherin was upregulated indicating that the process of
epithelial–mesenchymal transition (EMT) was restrained.
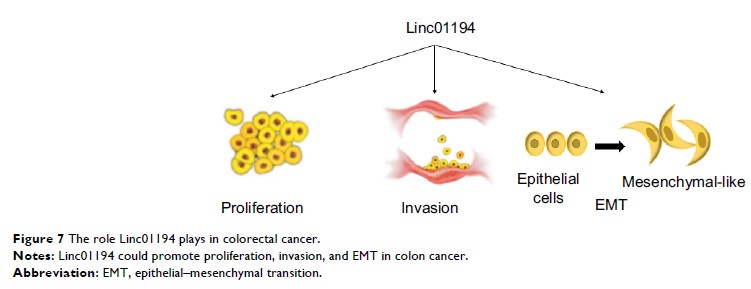
Conclusion: Linc01194 promotes
the proliferation and migration ability of colon cancer cells by activating
EMT. It acts as an oncogene in colorectal carcinoma and is associated with
worse survival outcome.
Keywords: colorectal
cancer, TCGA, lncRNA, Linc01194, EMT