108384
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

来自胃肝样腺癌的 GC-030-35 细胞系的建立和表征
Authors Wei J, Xue Y, Huo X, Han R, Su X, Jin Y, Zhao W, Chen Y, Zhang H, Dai J, Chen J
Received 4 September 2018
Accepted for publication 10 January 2019
Published 8 February 2019 Volume 2019:11 Pages 1275—1287
DOI https://doi.org/10.2147/CMAR.S186416
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Cristina Weinberg
Peer reviewer comments 1
Editor who approved publication: Dr Beicheng Sun
Purpose: Gastric hepatoid adenocarcinoma is a rare
subtype of primary gastric cancer and is a high-grade form of malignancy.
However, the pathogenesis and molecular biology of gastric hepatoid
adenocarcinoma remain poorly understood. The aim of this study was to establish
and characterize a new human gastric hepatoid adenocarcinoma cell line,
GC-030-35.
Materials and methods: The GC-030-35 cell line was established from tumor cells from a
58-year-old Chinese man with gastric hepatoid adenocarcinoma. The cultured
cells underwent immunocytochemistry and flow cytometry to confirm the tumor
cell phenotype. RNA sequencing was performed to analyze the differences in gene
expression between GC-030-35 cells compared with normal gastric epithelial
cells. A zebrafish assay was performed. Gene enrichment analysis and
interrogation of the bioinformatics databases, the Gene Ontology (GO) database
and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, were used for
pathway analysis.
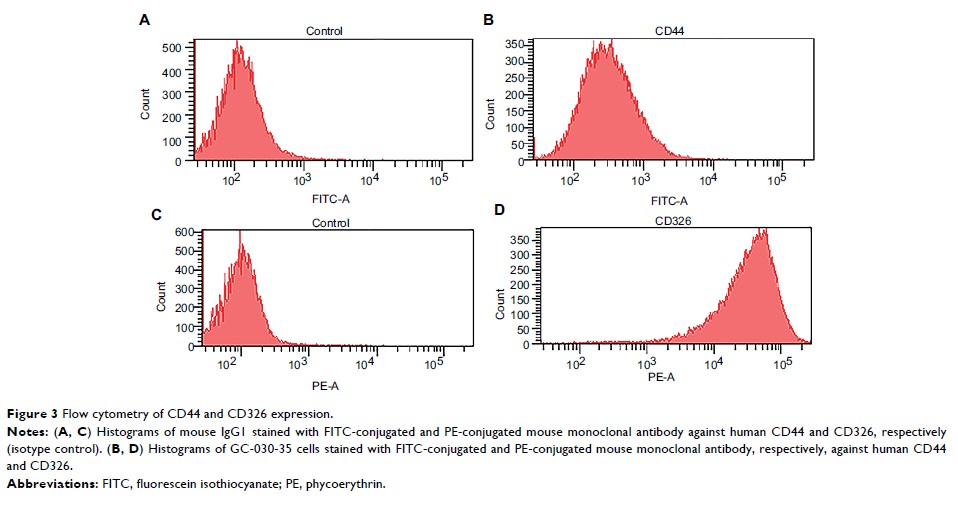
Results: Flow
cytometry analysis of the GC-030-35 cells showed a positive expression rate for
CD44+ of 10.7%, high cell clonality, an average plating efficiency of 32%,
cell-doubling time of 29.2 hours, and cell proliferation for >15 generations
in serial culture. The zebrafish assay showed the ability of the GC-030-35
cells to proliferate, promote angiogenesis, and metastasize. RNA sequencing
identified the functional clustering of 6,601 differentially expressed genes of
GC-030-35, which were significantly different when compared with nonneoplastic
gastric epithelial cells. Pathway enrichment analysis and interrogation of the
GO and KEGG bioinformatics databases identified genes for microbial metabolism
in diverse environments (63 genes), metabolism of xenobiotics by cytochrome P450
(CYP450; 25 genes), and the drug metabolism cytochrome P450 (28 genes).
Conclusion: A
human gastric hepatoid adenocarcinoma cell line, GC-030-35, was developed and
characterized by comparison with normal gastric epithelial cells.
Bioinformatics and gene analysis data showed that the CYP450 gene was
significantly differentially expressed by GC-030-35 cells.
Keywords: gastric
hepatoid adenocarcinoma, cell line, differentially expressed genes,
bioinformatics, carcinogenesis, cytochrome P450