108384
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

确定人类 2 型糖尿病胰岛的关键基因通路和共表达网络
Authors Li L, Pan Z, Yang S, Shan W, Yang Y
Received 2 July 2018
Accepted for publication 26 July 2018
Published 28 September 2018 Volume 2018:11 Pages 553—563
DOI https://doi.org/10.2147/DMSO.S178894
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Colin Mak
Peer reviewer comments 3
Editor who approved publication: Professor Ming-Hui Zou
Purpose: The number of people with type 2 diabetes (T2D) is growing rapidly worldwide. Islet β-cell dysfunction and failure are the main causes of T2D pathological processes. The aim of this study was to elucidate the underlying pathways and coexpression networks in T2D islets.
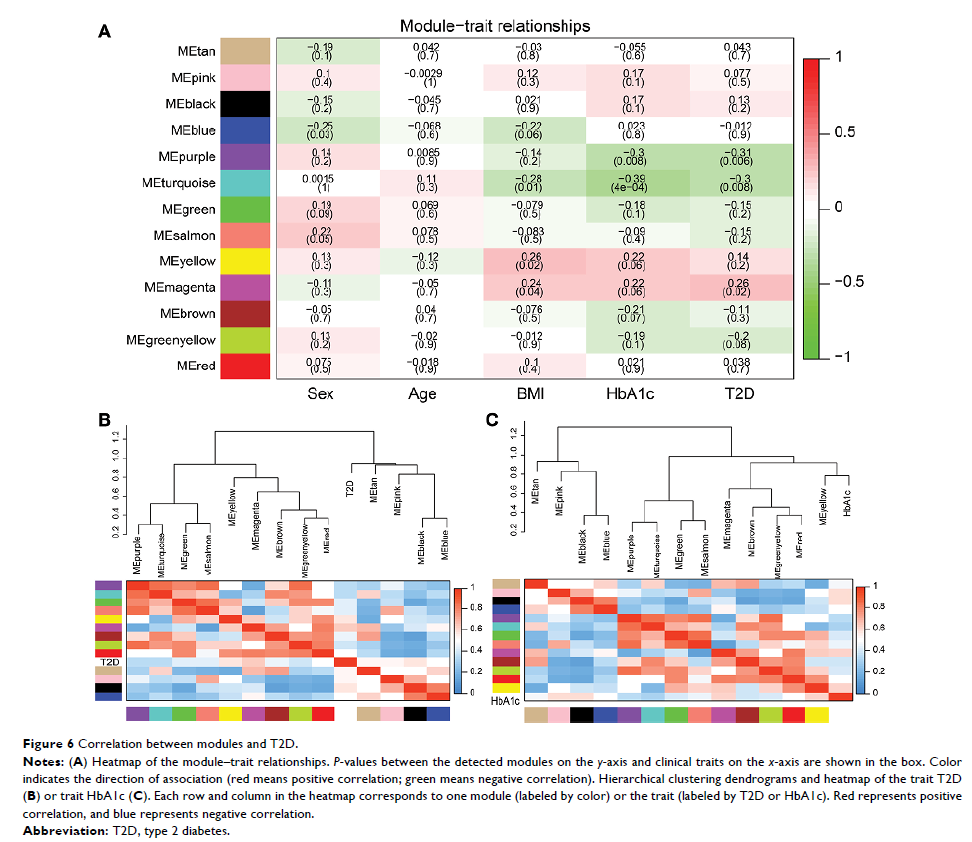
Materials and methods: We analyzed the differentially expressed genes (DEGs) in the data set GSE41762, which contained 57 nondiabetic and 20 diabetic samples, and developed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Protein–protein interaction (PPI) network, the modules from the PPI network, and the gene annotation enrichment of modules were analyzed as well. Moreover, a weighted correlation network analysis (WGCNA) was applied to screen critical gene modules and coexpression networks and explore the biological significance.
Results: We filtered 957 DEGs in T2D islets. Then GO and KEGG analyses identified that key pathways like inflammatory response, type B pancreatic cell differentiation, and calcium ion-dependent exocytosis were involved in human T2D. Three significant modules were filtered from the PPI network. Ribosome biogenesis, extrinsic apoptotic signaling pathway, and membrane depolarization during action potential were associated with the modules, respectively. Furthermore, coexpression network analysis by WGCNA identified 13 distinct gene modules of T2D islets and revealed four modules, which were strongly correlated with T2D and T2D biomarker hemoglobin A1c (HbA1c). Functional annotation showed that these modules mainly enriched KEGG pathways such as NF-kappa B signaling pathway, tumor necrosis factor signaling pathway, cyclic adenosine monophosphate signaling pathway, and peroxisome proliferators-activated receptor signaling pathway.
Conclusion: The results provide potential gene pathways and underlying molecular mechanisms for the prevention, diagnosis, and treatment of T2D.
Keywords: type 2 diabetes, islet β cell, bioinformatics analysis, differentially expressed genes, WGCNA