108384
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

通过全基因组测序分析确定 HER2 阳性和 HER2 阴性胃癌分子改变的差异
Authors Zhou C, Feng X, Yuan F, Ji J, Shi M, Yu Y, Zhu Z, Zhang J
Received 30 April 2018
Accepted for publication 24 July 2018
Published 26 September 2018 Volume 2018:10 Pages 3945—3954
DOI https://doi.org/10.2147/CMAR.S172710
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Colin Mak
Peer reviewer comments 3
Editor who approved publication: Professor Nakshatri
Objective: The aim of this study was to compare the molecular profiling, including somatic mutation and somatic copy number variation (SCNV), between human epidermal growth factor receptor 2 (HER2)-positive (HER2+) and HER2-negative (HER2–) gastric cancer patients.
Patients and methods: Tumor samples were collected from 15 gastric cancer patients, including 10 HER2+ samples and five HER2– samples, which were diagnosed by immunohistochemistry. Whole-genome sequencing was performed by Illumina HiSeq PE150 instrument, along with somatic single nucleotide variant (SNV), somatic structural variation (SV) and SCNV analyses.
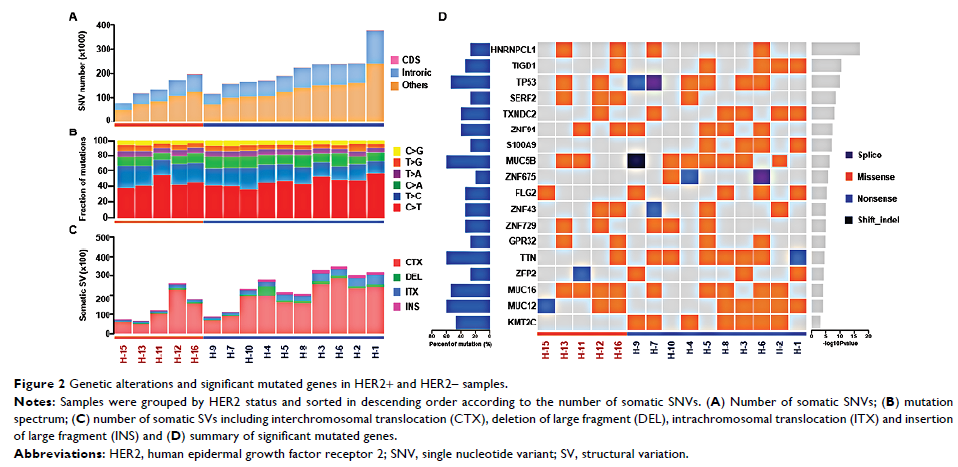
Results: The average number of somatic SNVs and mutation spectrum were similar between HER2+ and HER2– samples. Transition of C>T was the main type of mutation. For somatic SV, number of intrachromosomal translocation (2,850.3±1,260.4 vs 1,157±586.6, P =0.015) and insertion of large fragment (1,125.6±457.4 vs 500±138.9, P =0.002) in HER2+ samples were higher than those in HER2– samples. For all samples, lysine methyltransferase 2C (KMT2C ), ZNF91 , TAF1 and MAP4 genes were identified as new significant mutated driver genes. KMT2C gene mutations were mainly detected in HER2+ samples (7/10), which were correlated with the lysine degradation pathway. SERF2 gene mutations were more common in HER2– samples (3/5) than in HER2+ samples (1/10). Copy number gain was the major type of SCNV in both groups, and the average number of SCNVs was similar. In the HER2+ samples, by using the GISTIC algorithm, amplification of known driver genes cyclin-dependent kinase 12 (CDK12 , 6/10) and RARA (5/10) was mainly observed, and other amplifications including JUP , GJD3 , KRT39 , CDC6 , RAPGEFL1 , WIPF2 , FAM65C , KLF5 , DACH1 and PIBF1 genes were also observed. Amplifications of solute carrier family 12 member 7 (SLC12A7 , 5/5), TTC40 (4/5) and GALNT9 (4/5) genes were mainly detected in HER2– samples.
Conclusion: Differences in genomic landscape between HER2+ and HER2– gastric cancer samples were revealed in this study. KMT2C mutation and CDK12 amplification were mainly detected in HER2+ gastric cancer, whereas SERF2 mutation and SLC12A7 amplification were detected in HER2– gastric cancer.
Keywords: gastric cancer, HER2, whole-genome sequencing, gene mutation, gene amplification