108605
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

通过共表达网络分析确定 CDH11 与胃癌的进展和预后相关
Authors Chen PF, Wang F, Nie JY, Feng JR, Liu J, Zhou R, Wang HL, Zhao Q
Received 6 June 2018
Accepted for publication 10 August 2018
Published 2 October 2018 Volume 2018:11 Pages 6425—6436
DOI https://doi.org/10.2147/OTT.S176511
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Cristina Weinberg
Peer reviewer comments 3
Editor who approved publication: Dr Takuya Aoki
Background and
aims: Gastric cancer (GC) is one of the most
common cancers worldwide, and its pathogenesis is related to a complex network
of gene interactions. The aims of our study were to find hub genes associated
with the progression and prognosis of GC and illustrate the underlying
mechanisms.
Methods: Weighted gene co-expression network analysis (WGCNA)
was conducted using the microarray dataset and clinical data of GC patients
from Gene Expression Omnibus (GEO) database to identify significant gene modules
and hub genes associated with TNM stage in GC. Functional enrichment analysis
and protein–protein interaction network analysis were performed using the
significant module genes. We regarded the common hub genes in the co-expression
network and protein–protein interaction (PPI) network as “real” hub genes for
further analysis. Hub gene was validated in another independent dataset and The
Cancer Genome Atlas (TCGA) dataset.
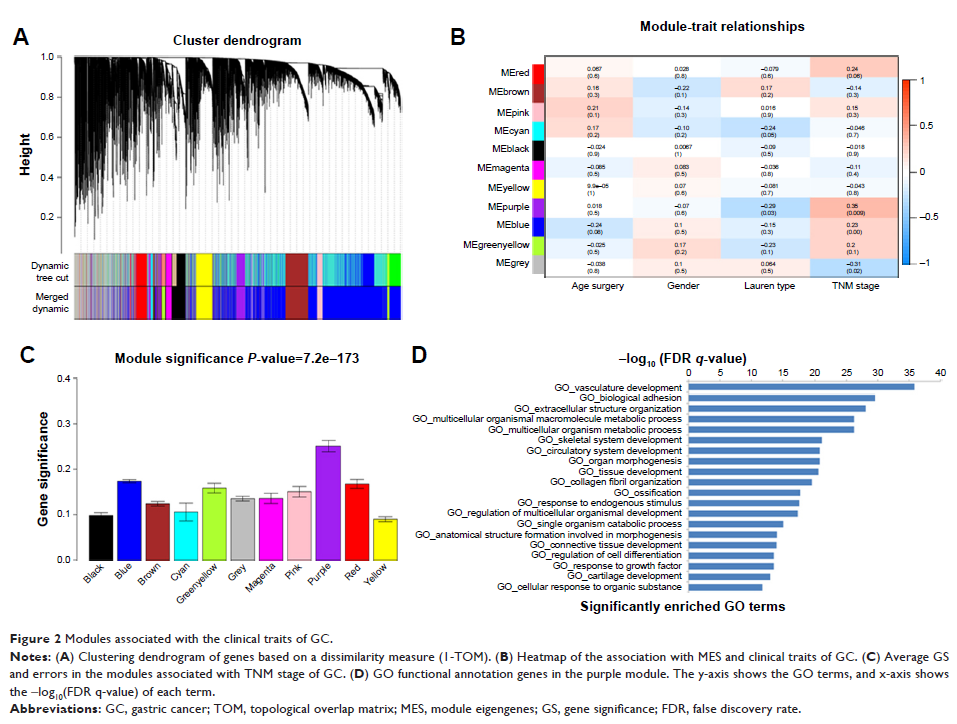
Results: In the significant purple module (R 2=0.35), a total
of 12 network hub genes were identified, among which six were also hub nodes in
the PPI network of the module genes. Functional annotation revealed that the
genes in the purple module focused on the biological processes of system
development, biological adhesion, extracellular structure organization and
metabolic process. In terms of validation, CDH11 had a higher correlation with
the TNM stage than other hub genes and was strongly correlated with biological
adhesion based on GO functional enrichment analysis. Data obtained from the
Gene Expression Profiling Interactive Analysis (GEPIA) showed that CDH11
expression had a strong positive correlation with GC stages (P <0.0001). In the testing set
and Oncomine dataset, CDH11 was highly expressed in GC tissues (P <0.0001). Survival analysis
indicated that samples with a high CDH11 expression showed a poor prognosis.
Cox regression analysis demonstrated an independent predictor of CDH11
expression in GC prognosis (HR=1.482, 95% CI: 1.015–2.164). Furthermore, gene
set enrichment analysis (GSEA) demonstrated that multiple tumor-related
pathways, especially focal adhesion, were enriched in CDH11 highly expressed
samples.
Conclusion: CDH11 was identified and validated in
association with progression and prognosis in GC, probably by regulating
biological adhesion and focal adhesion-related pathways.
Keywords: gastric cancer,
weighted gene co-expression network, hub gene, prognosis