108384
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

分析与膀胱癌原发性纤毛相关的潜在基因
Authors Du E, Lu C, Sheng F, Li C, Li H, Ding N, Chen Y, Zhang T, Yang K, Xu Y
Received 26 May 2018
Accepted for publication 21 June 2018
Published 29 August 2018 Volume 2018:10 Pages 3047—3056
DOI https://doi.org/10.2147/CMAR.S175419
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Justinn Cochran
Peer reviewer comments 2
Editor who approved publication: Professor Nakshatri
Background: Dysfunction of primary cilia (PC), which could influence cell cycle and modulate cilia-related signaling transduction, has been reported in several cancers. However, there is no evidence of their function in bladder cancer (BLCA). This study was performed to investigate the presence of PC in BLCA and to explore the potential molecular mechanisms underlying the PC in BLCA.
Patients and methods: The presence of PC was assessed in BLCA and adjacent non-cancerous tissues. The gene expression dataset GSE52519 was employed to obtain differentially expressed genes (DEGs) associated with PC. The mRNA expression of the DEGs were confirmed by Gene Expression Profiling Interactive Analysis. The DEGs properties and pathways were analyzed by Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis. Genomatix software was used to predict putative transcription factor binding sites (TFBS) in the promoter region of DEGs, and the transcription factors were achieved according to the shared TFBS, which were supported by the ChIP-Sequence data.
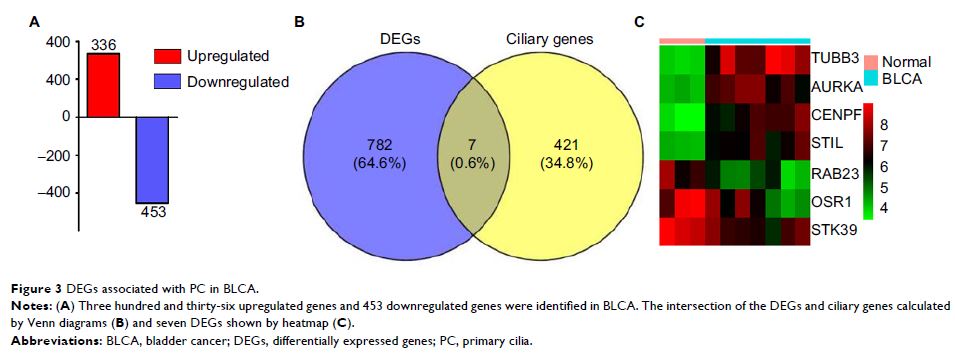
Results: PC were found to be reduced in BLCA tissue samples in this study. Seven DEGs were observed to be associated with PC, and gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis indicated that these DEGs exhibited the properties and functions of PC, and that the Hedgehog signaling pathway probably participated in the pathogenesis and progression of BLCA. The mRNA expression of the seven DEGs in 404 BLCA and 28 normal tissue samples were analyzed, and five DEGs including CENPF , STIL , AURKA , STK39 and OSR1 were identified. Five TFBS including CREB, E2FF, EBOX, ETSF and HOXF in the promoter region of five DEGs were calculated and the transcription factors were obtained according to the shared TFBS.
Conclusion: PC were found to be reduced in BLCA, and the potential molecular mechanisms of PC in BLCA helped to provide novel diagnosis and therapeutic targets for BLCA.
Keywords: bladder cancer, primary cilia, differentially expressed genes, microarray analysis, transcription factors