108899
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

通过蛋白质组学和代谢组学测试严重抑郁症患者磷脂代谢的血浆紊乱
Authors Gui SW, Liu YY, Zhong XG, Liu XY, Zheng P, Pu JC, Zhou J, Chen JJ, Zhao LB, Liu LX, Xu GW, Xie P
Received 30 January 2018
Accepted for publication 6 April 2018
Published 6 June 2018 Volume 2018:14 Pages 1451—1461
DOI https://doi.org/10.2147/NDT.S164134
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Amy Norman
Peer reviewer comments 3
Editor who approved publication: Professor Wai Kwong Tang
Introduction: Major
depressive disorder (MDD) is a highly prevalent mental disorder affecting
millions of people worldwide. However, a clear causative etiology of MDD
remains unknown. In this study, we aimed to identify critical protein
alterations in plasma from patients with MDD and integrate our proteomics and
previous metabolomics data to reveal significantly perturbed pathways in MDD.
An isobaric tag for relative and absolute quantification (iTRAQ)-based
quantitative proteomics approach was conducted to compare plasma protein
expression between patients with depression and healthy controls (CON).
Methods: For integrative analysis, Ingenuity Pathway Analysis software was
used to analyze proteomics and metabolomics data and identify potential
relationships among the differential proteins and metabolites.
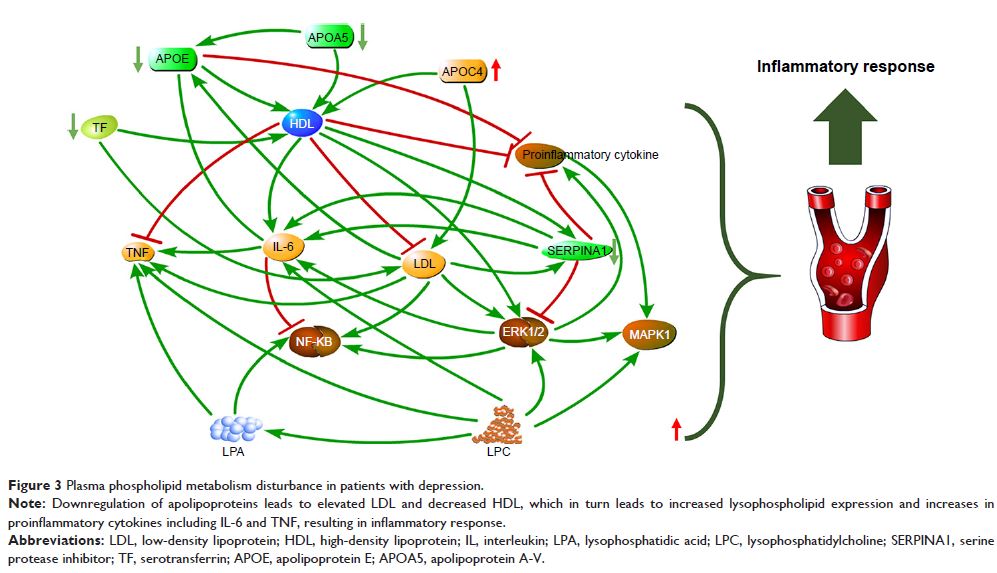
Results: A total of 74 proteins were significantly changed in patients with
depression compared with those in healthy CON. Bioinformatics analysis of
differential proteins revealed significant alterations in lipid transport and
metabolic function, including apolipoproteins (APOE, APOC4 and APOA5), and the
serine protease inhibitor. According to canonical pathway analysis, the top
five statistically significant pathways were related to lipid transport,
inflammation and immunity.
Conclusion: Causal network analysis by integrating differential proteins and metabolites
suggested that the disturbance of phospholipid metabolism might promote the
inflammation in the central nervous system.
Keywords: major depressive disorder, plasma proteomics, iTRAQ, metabolomics,
integrative analysis