108985
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

上调 VEGFA 和 DLL4 可成为肾透明细胞癌的潜在预后基因
Authors Wang X, Zhang J, Wang Y, Tu M, Wang Y, Shi G
Received 1 September 2017
Accepted for publication 8 December 2017
Published 26 March 2018 Volume 2018:11 Pages 1697—1706
DOI https://doi.org/10.2147/OTT.S150565
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Akshita Wason
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Purpose: As a typical hypervascular tumor, clear cell renal cell carcinoma
(ccRCC) is the most common type of RCC. This study was aimed to explore the
prognostic genes for ccRCC, focusing on the roles of vascular endothelial
growth factor A (VEGFA ) and Delta-like ligand 4 (DLL4 ) in the disease.
Materials and
methods: The mRNA-sequencing data of kidney
renal clear cell carcinoma (KIRC) were obtained from The Cancer Genome Atlas
(TCGA) database, including 469 tumor samples and 68 adjacent normal samples.
Using limma package, differentially expressed genes (DEGs) were analyzed by
differential expression and subgroup analyses and confirmed using validation
dataset GSE53757. Followed by enrichment analysis, protein–protein interaction
(PPI) network analysis and protein subcellular localization were performed
using multifaceted analysis tool for human transcriptome tool, and Cytoscape
software and InnateDB database, respectively. Moreover, survival analysis was
conducted to identify key prognosis-associated genes. In addition, VEGFA and DLL4 levels were detected
using real-time quantitative PCR (qRT-PCR).
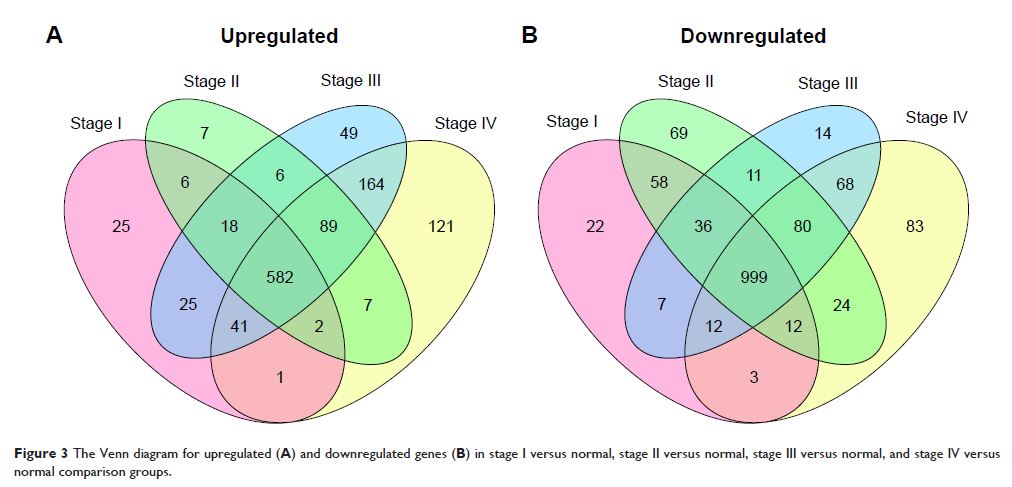
Results: A total of 1,984 DEGs were screened in the KIRC tumor
samples. VEGFA was located in
extracellular space and could interact with placental growth factor (PGF) and
angiopoietin 2 (ANGPT2) in the PPI network. Subgroup analysis suggested
that VEGFA was significantly
upregulated in stages I, II, and III ccRCC tumor samples. Survival analysis
showed that TIMP1 was among the top four
prognosis-associated genes. qRT-PCR analysis confirmed that the expression
levels of DLL4 and VEGFA were significantly
upregulated in tumor samples.
Conclusion: VEGFA and DLL4 might be
prognostic genes for ccRCC. Besides, PGF , ANGPT2 , and TIMP1 might also be related
to the prognosis of ccRCC patients.
Keywords: clear cell renal cell carcinoma, differentially expressed genes,
protein–protein interaction network, subgroup analysis, survival analysis