108985
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

上调的长非编码 RNA 对乳腺癌检测有良好的功效: 一项综合分析
Authors Yu G, Zhang W, Zhu L, Xia L
Received 21 September 2017
Accepted for publication 28 November 2017
Published 16 March 2018 Volume 2018:11 Pages 1491—1499
DOI https://doi.org/10.2147/OTT.S152241
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Akshita Wason
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Purpose: Focusing on the latest literature, dysregulated long non-coding RNAs
(lncRNAs) have been extensively explored in breast cancer (BC) research. The
purpose of this meta-analysis is to synthesize the evidence on the diagnostic
performance of abnormally expressed lncRNAs for BC.
Materials and
methods: Relevant studies were searched
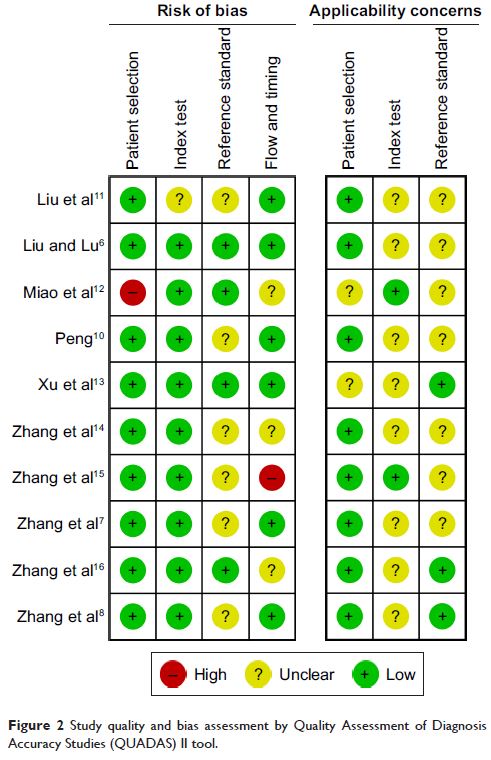
in multiple electronic databases. The Quality Assessment of Diagnosis Accuracy
Studies II criteria were applied to assess the quality of included studies. The
bivariate meta-analysis model was applied to synthesize the diagnostic
parameters using Stata 12.0 software. Publication bias was judged in terms of
the Deek’s funnel plot asymmetry test.
Results: We included 10 eligible studies, which comprised 835 BC patients
and 725 paired controls for this meta-analysis. The pooled sensitivity,
specificity, diagnostic odds ratio, likelihood ratio positive, likelihood ratio
negative, and area under the curve (AUC) of upregulated lncRNA expression
signature in confirming BC were 0.79 (95% CI: 0.70–0.85), 0.80 (95% CI:
0.73–0.85), 14.61 (95% CI: 10.91–19.55), 3.90 (95% CI: 3.03–5.02), 0.27 (95%
CI: 0.20–0.36), and 0.86, respectively. Stratified analyses yielded a
sensitivity of 0.83 (95% CI: 0.80–0.86) for serum-based analysis, which was
higher than plasma-based analysis, whereas plasma-based analysis revealed a
greater specificity of 0.88 (95% CI: 0.85–0.91). Moreover, lncRNA-homeotic
genes (HOX) transcript antisense RNA showed a pooled specificity of 0.89 (95%
CI: 0.84–0.93) and AUC of 0.86, which were superior to performances by
lncRNA-metastasis-associated lung adenocarcinoma transcript-1 and -H19 in
diagnosing BC. Notably, the analysis based on cancer subtypes demonstrated that
lncRNA expression signature could distinguish triple-negative BC (lacks
estrogen receptor, progesterone receptor, and human epidermal growth factor
receptor 2 expression) from non-triple-negative BC, with an AUC of 0.85.
Conclusion: Upregulated lncRNAs reveal an immense potential as novel
non-invasive biomarker(s) that could complement BC diagnosis.
Keywords: lncRNA, breast cancer, diagnosis, meta-analysis