108985
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

三阴性乳腺癌中与 cRNA 对话网络(crosstalk network)相关联的带有编码 RNA 的 lncRNAs 和 miRNA 的整合分析
Authors Yuan NJ, Zhang G, Bie FJ, Ma M, Ma Y, Jiang X, Wang Y, Hao X
Received 16 August 2017
Accepted for publication 26 September 2017
Published 12 December 2017 Volume 2017:10 Pages 5883—5897
DOI https://doi.org/10.2147/OTT.S149308
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Amy Norman
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Abstract: Triple negative breast cancer (TNBC) is a particular subtype of breast
malignant tumor with poorer prognosis than other molecular subtypes. Currently,
there is increasing focus on long non-coding RNAs (lncRNAs), which can act as
competing endogenous RNAs (ceRNAs) and suppress miRNA functions involved in
post-transcriptional regulatory networks in the tumor. Therefore, to
investigate specific mechanisms of TNBC carcinogenesis and improve treatment
efficiency, we comprehensively integrated expression profiles, including data
on mRNAs, lncRNAs and miRNAs obtained from 116 TNBC tissues and 11 normal
tissues from The Cancer Genome Atlas. As a result, we selected the threshold
with |log2FC|>2.0 and an adjusted p-value >0.05 to
obtain the differentially expressed mRNAs, miRNAs and lncRNAs. Hereafter,
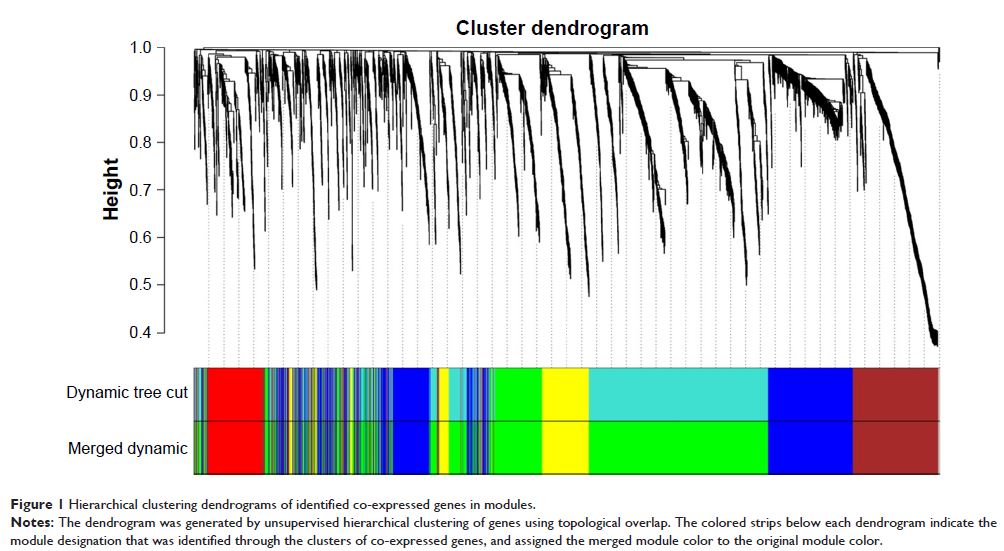
weighted gene co-expression network analysis was performed to identify the
expression characteristics of dysregulated genes. We obtained five
co-expression modules and related clinical feature. By means of correlating
gene modules with protein–protein interaction network analysis that had
identified 22 hub mRNAs which could as hub target genes. Eleven key
dysregulated differentially expressed micro RNAs (DEmiRNAs) were identified
that were significantly associated with the 22 hub potential target genes.
Moreover, we found that 14 key differentially expressed lncRNAs could interact
with the key DEmiRNAs. Then, the ceRNA crosstalk network of TNBC was
constructed by utilizing key lncRNAs, key miRNAs, and hub mRNAs in Cytoscape
software. We analyzed and described the potential characteristics of biological
function and pathological roles of the TNBC ceRNA co-regulatory network; also,
the survival analysis was performed for each molecule. These findings revealed
that ceRNA crosstalk network could play an important role in the development
and progression for TNBC. In addition, we also identified that some molecules
in the ceRNA network possess clinical correlation and prognosis.
Keywords: triple negative breast cancer, The Cancer Genome Atlas, weighted
gene co-expression network analysis, ceRNA crosstalk network, survival
prognosis