108985
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

CYP3A4 变异体对利多卡因体外代谢能力的功能评估
Authors Fang P, Tang P, Xu R, Zheng X, Wen J, Bao S, Cai J, Hu G
Received 22 September 2017
Accepted for publication 25 October 2017
Published 7 December 2017 Volume 2017:11 Pages 3503—3510
DOI https://doi.org/10.2147/DDDT.S152366
Checked for plagiarism Yes
Review by Single-blind
Peer reviewers approved by Dr Akshita Wason
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Aim: Human cytochrome P450 3A4 is the most abundant isoform of P450
enzyme in the liver. It plays an important role in the metabolism of wide
variety of xenobiotic and endogenous substrates. So far, there are few reports
about the functional characterization of CYP3A4 variants
in terms of specific substrates. The aim of this study was to systematically
investigate the genetic polymorphisms of 23 CYP3A4 alleles
and evaluate their catalytic activities on the metabolism of lidocaine in
vitro.
Methods and results: The wild-type and 22 CYP3A4 variants
were expressed in Spodoptera frugiperda 21 insect cells. Then the insect
microsomes were incubated with the CYP3A4-specific substrate lidocaine.
Reactions were performed with 50–3,000 µM for 60 min at 37°C. Lidocaine and its
metabolite monoethylglycinexylidide were analyzed by ultra-performance liquid
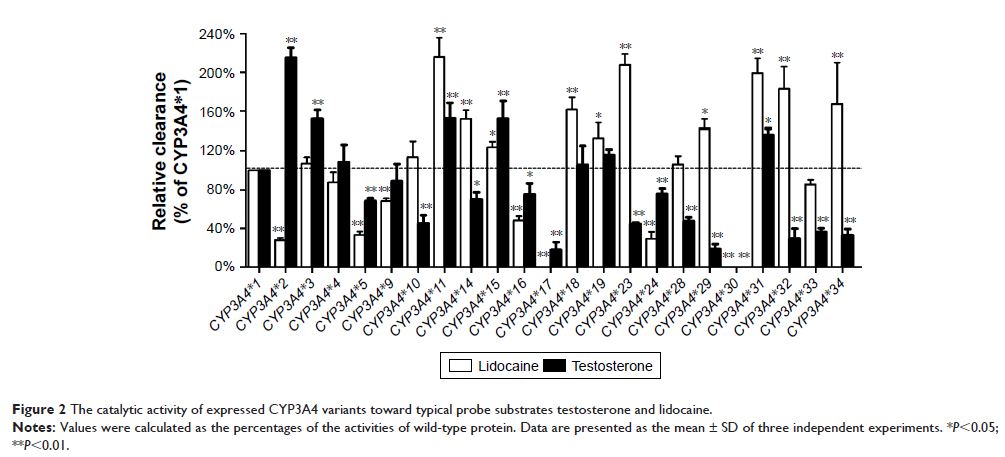
chromatography-tandem mass spectrometry system. Of the 23 CYP3A4 allelic variants
tested, 2 variants (CYP3A4 *17 and CYP3A4 *30 )
had no detectable enzyme activity; and 5 variants (CYP3A4 *2 , CYP3A4 *5 , CYP3A4 *9 , CYP3A4 *16 and CYP3A4 *24 )
showed significantly decreased intrinsic clearance values compared with
wild-type CYP3A4* 1.
Conclusion: As the first study of all these CYP3A4 alleles for lidocaine
metabolism, our results in vitro assessment may provide novel insights into the
allele-specific and substrate-specific activity of CYP3A4 and may also offer a
reference to the personalized treatment of lidocaine in a clinical setting.
Keywords: CYP3A4 , genetic polymorphism, drug
metabolism, lidocaine, personalized treatment