108605
论文已发表
注册即可获取德孚的最新动态
IF 收录期刊
- 3.4 Breast Cancer (Dove Med Press)
- 3.2 Clin Epidemiol
- 2.6 Cancer Manag Res
- 2.9 Infect Drug Resist
- 3.7 Clin Interv Aging
- 5.1 Drug Des Dev Ther
- 3.1 Int J Chronic Obstr
- 6.6 Int J Nanomed
- 2.6 Int J Women's Health
- 2.9 Neuropsych Dis Treat
- 2.8 OncoTargets Ther
- 2.0 Patient Prefer Adher
- 2.2 Ther Clin Risk Manag
- 2.5 J Pain Res
- 3.0 Diabet Metab Synd Ob
- 3.2 Psychol Res Behav Ma
- 3.4 Nat Sci Sleep
- 1.8 Pharmgenomics Pers Med
- 2.0 Risk Manag Healthc Policy
- 4.1 J Inflamm Res
- 2.0 Int J Gen Med
- 3.4 J Hepatocell Carcinoma
- 3.0 J Asthma Allergy
- 2.2 Clin Cosmet Investig Dermatol
- 2.4 J Multidiscip Healthc

已发表论文
宫颈鳞癌中的替代性拼接景观
Authors Guo P, Wang D, Wu J, Yang J, Ren T, Zhu B, Xiang Y
Published Date December 2014 Volume 2015:8 Pages 73—79
DOI http://dx.doi.org/10.2147/OTT.S72832
Received 18 August 2014, Accepted 13 November 2014, Published 22 December 2014
Background: Alternative splicing (AS) is a key regulatory mechanism in protein
synthesis and proteome diversity. In this study, we identified alternative
splicing events in four pairs of cervical squamous cell carcinoma (CSCC) and
adjacent nontumor tissues using RNA sequencing.
Methods: The transcripts of the four paired samples were thoroughly analyzed by RNA sequencing. SpliceMap software was used to detect the splicing junctions. Kyoto Encyclopedia of Genes and Genomes pathway analysis was conducted to detect the alternative spliced genes-related signal pathways. The alternative spliced genes were validated by reverse transcription-polymerase chain reaction (RT-PCR).
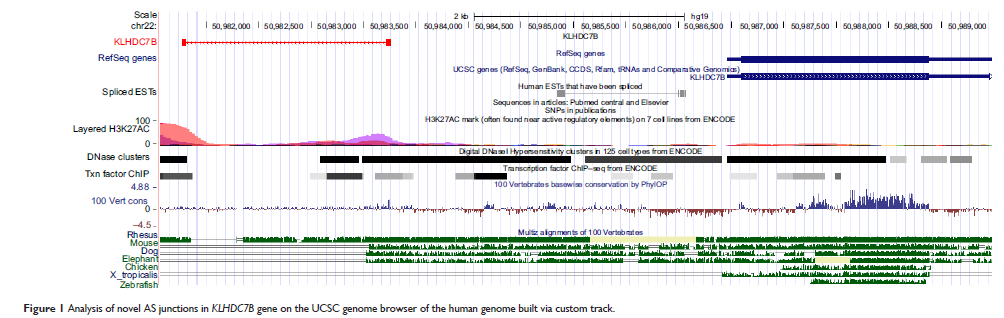
Results: There were 35 common alternative spliced genes in the four CSCC samples; they were novel and CSCC specific. Sixteen pathways were significantly enriched (P <0.05). One novel 5'AS site in the KLHDC7B gene, encoding kelch domain-containing 7B, and an exon-skipping site in the SYCP2 gene, encoding synaptonemal complex 2, were validated by RT-PCR. The KLHDC7B gene with 5'AS was found in 67.5% (27/40) of CSCC samples and was significantly related with cellular differentiation and tumor size. The exon-skipping site of the SYCP2 gene was found in 35.0% (14/40) of CSCC samples and was significantly related with depth of cervical invasion.
Conclusion: The KLHDC7B and the SYCP2 genes with alternative spliced events might be involved in the development and progression of CSCC and could be used as biomarkers in the diagnosis and prognosis of CSCC.
Keywords: cervical squamous carcinomas, alternative splicing events, RNA sequencing
Methods: The transcripts of the four paired samples were thoroughly analyzed by RNA sequencing. SpliceMap software was used to detect the splicing junctions. Kyoto Encyclopedia of Genes and Genomes pathway analysis was conducted to detect the alternative spliced genes-related signal pathways. The alternative spliced genes were validated by reverse transcription-polymerase chain reaction (RT-PCR).
Results: There were 35 common alternative spliced genes in the four CSCC samples; they were novel and CSCC specific. Sixteen pathways were significantly enriched (P <0.05). One novel 5'AS site in the KLHDC7B gene, encoding kelch domain-containing 7B, and an exon-skipping site in the SYCP2 gene, encoding synaptonemal complex 2, were validated by RT-PCR. The KLHDC7B gene with 5'AS was found in 67.5% (27/40) of CSCC samples and was significantly related with cellular differentiation and tumor size. The exon-skipping site of the SYCP2 gene was found in 35.0% (14/40) of CSCC samples and was significantly related with depth of cervical invasion.
Conclusion: The KLHDC7B and the SYCP2 genes with alternative spliced events might be involved in the development and progression of CSCC and could be used as biomarkers in the diagnosis and prognosis of CSCC.
Keywords: cervical squamous carcinomas, alternative splicing events, RNA sequencing